Biomarker Discovery for Alzheimer’s Disease
By Jon Armstrong
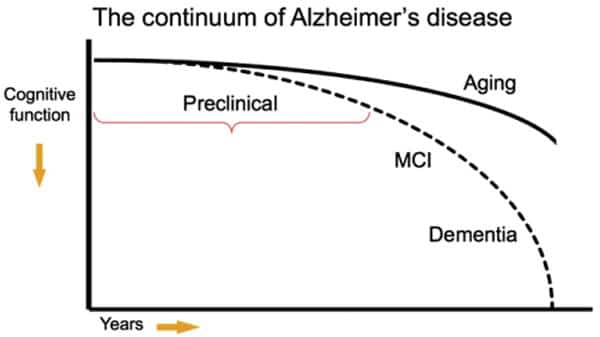
As the leader of R&D for Cofactor, I am of course laser focused on current work, while also thinking about how our technology might be applied to diseases that are not in our current stack. One of these diseases is Alzheimer’s Disease (AD). AD, being a disease of the brain, presents unique challenges in generating diagnostic, prognostic and predictive biomarkers from the direct site of disease, since brain biopsies aren’t possible. Current methods for diagnosis include a full clinical workup, including neurophysiological testing coupled with brain imaging using magnetic resonance imaging. However, even after testing, discriminating between AD and non-AD dementia using these non-biomarker based clinical diagnostic criteria results in around 32% of patients receiving an ambiguous or misdiagnosis of AD.¹ Thus, it is important to determine biomarkers that provide increase clinical diagnostic accuracy, yet even more challenges exist at earlier stages of the disease. However, it has been reported that biochemical changes occur up to two decades prior to the presentation of clinical symptoms² providing an opportunity for early and differential diagnosis of AD.
Diseases in the brain, such as AD, require one to move farther away from the site of disease to more distant compartments for discovery and diagnosis. A classic example in AD of one of these compartments is the cerebral spinal fluid (CSF). Much work has been focused on identifying protein biomarkers for AD in CSF. To date, this work has resulted in three core CSF protein biomarkers for AD diagnosis, the 42 amino acid long amyloid-beta peptide (Aβ1-42), total tau protein (T-tau), and tau phosphorylated at threonine 181 (P-tau181).³ However, the pathology of late-onset AD is heterogenous and there is a need for improved AD CSF biomarkers.
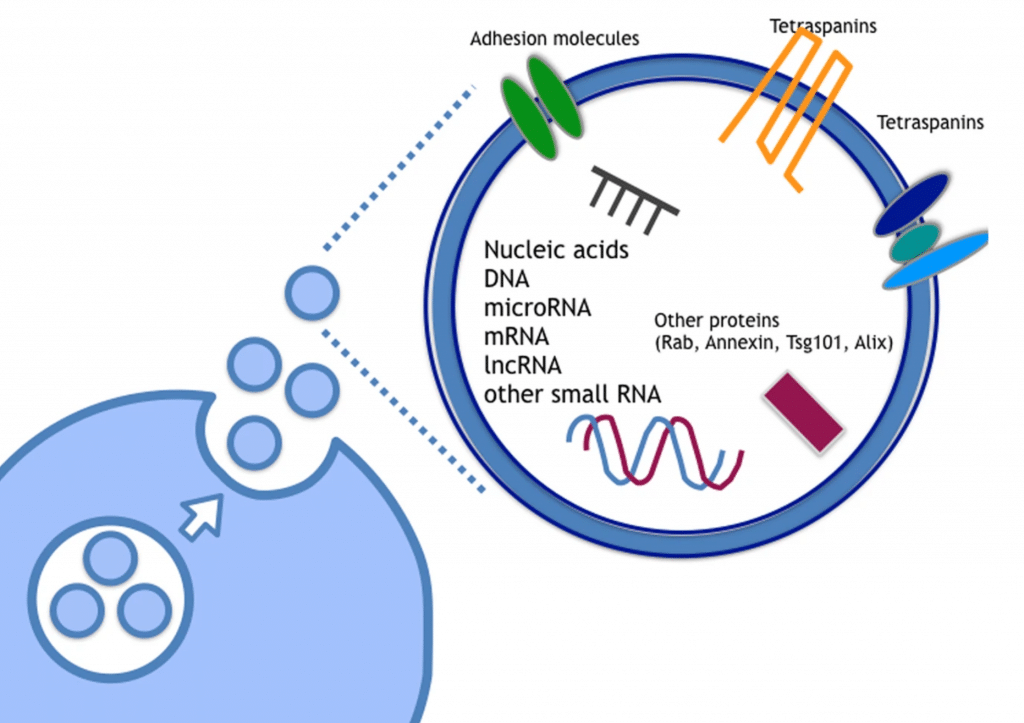
One area of promise for the detection of AD biomarkers from CSF or other fluids such as blood or plasma are extracellular vesicles (EVs). EVs are lipid-bilayer delimited particles that are naturally released from virtually all cell types. EVs have been shown to facilitate neuron-glia communication, promote neuronal progression and regeneration, and also contribute to the development of glioblastoma and neurodegenerative diseases.⁴⁻⁷ Of interest, EVs show relative amounts of heterogenous molecular species, such as nucleic acids, lipids, and proteins which may depend on their cellular origin.⁸ With a focus on nucleic acids, mRNA, long and short non-coding RNAs, microRNA, and mitochondrial DNA have been found in EVs with miRNA attracting early attention due to their role in regulating gene expression.⁹⁻¹¹ Recently, mRNA and long non-coding RNA have shown to be potential biomarkers for cancer.12,13 EVs continue to be an intriguing target for the discover of RNA biomarkers since RNAs in EVs are stable in the human body and protected from RNAses, which are abundant in human body fluids, since they are contained in the EV. This resistance affords several advantages to using RNAs from EVs as disease biomarkers, such as reproducibility, storage and sensitivity. However, to date, nearly all of the biomarker work with EVs has focused on single analyte biomarkers (e.g. MMP1 mRNA in ovarian cancer and the long non-coding RNA HOTTIP in gastric cancer). Considering the contents of EVs are complex and heterogenous and a disease such AD shows heterogeneity of pathology, it is likely that a more comprehensive and multidimensional biomarker strategy will be needed for early and accurate non-invasive diagnosis of AD.
Cofactor’s platform technology gives us the ability to assay the RNA from a specific sample and generate Health Expression Models using machine learning and advanced analytics. We can then extend those Health Expression Models to cohorts of patient samples, utilizing patient meta-data (e.g. treatment, time to recurrence, etc.), to further develop Predictive Models based on multi-dimensional biomarkers to identify treatment responders and non-responders. The current focus of our technology is in oncology, specifically immunocology, however one of the aspects of my job is to consider how our technology might be applied to other disease areas and sample types in the future. I find it exciting to think that in the future our technology might be applied to a disease like AD, where RNA is gathered, not directly from a cancer biopsy, but from EVs. The RNA signals that we generate from the EVs could then be analyzed using our platform technology and Health Expression Models could be generated to better diagnose AD. Even though heterogeneity exists in the RNA material contained in EVs and with the pathology of AD, our platform is designed to not look at single analyte biomarkers such as those mentioned above (which would likely suffer from material and pathological noise), but to consider all of the RNA signals in the EVs to distinguish the most sensitive and reliable multi-dimensional biomarkers for a Health Expression Model of AD. The obvious application of an AD Health Expression Model would be to develop a system for the early detection of AD. In closing, my hope is that with better detection and diagnosis comes the development of drugs for the treatment of AD. AD is a devastating disease which effects encompass more than just scientific challenges; it’s effects reach to our payer systems, healthcare and clinician networks, and most importantly, the emotional and personal cost to the families and friends of loved ones that are living or have died from AD.
- Bjerke M, Engelborghs S. Cerebrospinal Fluid Biomarkers for Early and Differential Alzheimer’s Disease Diagnosis. J Alzheimers Dis, 2018, 62:1199–209
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR, Kaye J, Montine TJ, Park DC, Reiman EM, Rowe CC, Siemers E, Stern Y, Yaffe K, Carrillo MC, Thies B, Morrison-Bogorad M, Wagster M V., Phelps CH. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement, 2011, 7:280–92
- Mattsson N, Zetterberg H, Blennow kaj. Lessons from Multicenter Studies on CSF Biomarkers for Alzheimer’s Disease. Int J Alzheimers Dis, 2010, 2010:1–5
- Janas AM, Sapoń K, Janas T, Stowell MHB, Janas T. Exosomes and other extracellular vesicles in neural cells and neurodegenerative diseases. Biochim Biophys Acta – Biomembr, 2016, 1858:1139–51
- Von Bartheld CS, Altick AL. Multivesicular bodies in neurons: Distribution, protein content, and trafficking functions. Prog Neurobiol, 2011, 93:313–40
- Bronisz A, Wang Y, Nowicki MO, Peruzzi P, Ansari KI, Ogawa D, Balaj L, De Rienzo G, Mineo M, Nakano I, Ostrowski MC, Hochberg F, Weissleder R, Lawler SE, Chiocca EA, Godlewski J. Extracellular Vesicles Modulate the Glioblastoma Microenvironment via a Tumor Suppression Signaling Network Directed by miR-1. Cancer Res, 2014, 74:738–50
- Surgucheva I, Sharov VS, Surguchov A. γ-Synuclein: Seeding of α-Synuclein Aggregation and Transmission between Cells. Biochemistry, 2012, 51:4743–54
- Mulcahy LA, Pink RC, Carter DRF. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles, 2014, 3:24641
- Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genomics Proteomics Bioinformatics, 2015, 13:17–24
- Vlassov A V., Magdaleno S, Setterquist R, Conrad R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta – Gen Subj, 2012, 1820:940–8
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol, 2007, 9:654–9
- Yokoi A, Yoshioka Y, Yamamoto Y, Ishikawa M, Ikeda S, Kato T, Kiyono T, Takeshita F, Kajiyama H, Kikkawa F, Ochiya T. Malignant extracellular vesicles carrying MMP1 mRNA facilitate peritoneal dissemination in ovarian cancer. Nat Commun, 2017, 8:14470
- Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X, Liu Y, Zhang Y. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer, 2018, 17:68