Solid Tumor Analysis in Cancer
By Danielle M. Quintanilha, PhD
Cancer is not only a deeply devastating disease for patients and their families, but is cited as the second leading cause of death (29%) in the country, second only to heart diseases. Cancer is a collective name of related genetic diseases, caused by oncogene activation and/or tumor suppressor gene inhibition. These genetic mutations lead to uncontrolled cell division and spread into healthy surrounding tissues. Cancers can be divided in two types: solid tumor cancers and blood cancers. A solid tumor is an abnormal mass of tissue that usually does not contain cysts or liquid areas. Solid tumor cancers (i.e. sarcomas, carcinomas, and lymphomas) can develop in many parts of the human body and account for over 90% of all human cancer cases in the United States. Beyond initial diagnosis of solid tumors through imaging techniques and tissue pathology analysis, molecular assays are available to provide further solid tumor analysis and characterize the cancer.
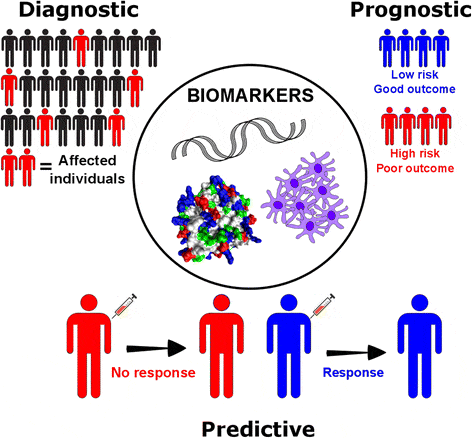
Molecular assays consist of analytical tests of measurable molecular traits commonly referred to as cancer biomarkers. These biomarkers can provide diagnostic, prognostic or predictive clinical information (figure 1). Diagnostic biomarkers can confirm the presence of the cancer, narrow down the specific genetic mutation diagnosis and sometimes enable early cancer detection. Prognostic biomarkers inform on the likely patient health outcome (independent of treatment administration) and can predict the risk of cancer recurrence. Predictive biomarkers can match the use of available targeted therapies, immune therapies or clinical trials to patients who could benefit the most from them. Biomarker molecular assays are performed directly on tumor cells, or by analyzing DNA, RNA or proteins procured from tumor tissue biopsies or liquid biopsies.
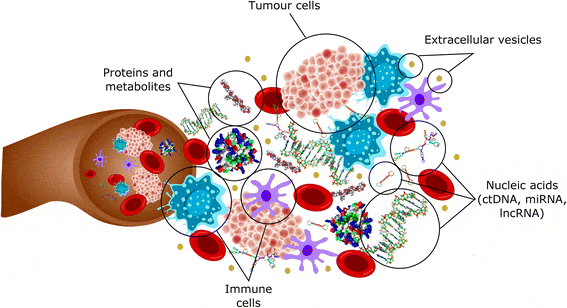
Solid tumor analysis, including molecular assays, can be categorized according to the specimen type collected prior to biomarker analysis. The specimen can be the tumor tissue itself obtained through fine needle, core needle or incisional biopsy, or it can be body fluids. Liquid biopsies are molecular assays that use collected peripheral blood, urine, saliva, pleural effusions, cerebrospinal fluid or stool samples. These samples are used to harvest circulating tumor cells (CTCs), circulating tumor nucleic acids (DNA and RNA) and proteins for biomarker detection. Figure 2 depicts circulating biomarkers that can be assessed with liquid biopsy assays.
Nowadays molecular testing is a standard strategy for solid tumor analysis and classifying solid tumors. They follow guidelines provided by the College of American Pathologists (CAP) and the National Comprehensive Cancer Network (NCCN). Some of the different molecular assays for solid tumor analysis are briefly described below:
1. Liquid Biopsy of Circulating Tumor Cells (CTCs): Circulating tumor cells are cancer cells that detach from a primary tumor and travel through the vascular system to other parts of the body. CTCs play an important role in metastasis formation and can be used for early cancer diagnosis and therapeutic decision making. The FDA has approved the immunoaffinity-based assay CellSearch CTC Test for detecting CTCs in metastatic breast, prostate and colorectal cancer. This quantitative assay discriminates CTCs using ferrofluid nanoparticles coupled with an antibody against epithelial cell adhesion molecule (EpCAM), which is highly expressed in tumor cells of epithelial origin. However, many CTCs can downregulate the expression of cell surface markers used for capture (e.g. through the process of epithelial-mesenchymal transition), posing a difficulty to the efficacy of this test.
2. Liquid Biopsy of Circulating Tumor DNA (ctDNA) and RNA: Tumor DNA and RNA are released into the bloodstream when tumor cells go through apoptosis, necrosis or exosome excretion. Molecular assays using ctDNA and RNA that combine polymerase chain reaction-based techniques and Next-Generation Sequencing can identify single nucleotide variants (SNV), small insertions/deletions (indel), copy number alterations (CNV) and fusion genes. Similar assays can also be performed using quantitative PCR or digital droplet PCR. The advantage of liquid biopsy assays is that the collection of the samples is typically less invasive than traditional biopsies, and for some patients a biopsy procedure is not a possibility. Limitations of these assays are related to the low frequency of mutant alleles in a high background of wild-type cell-free DNA/RNA as well as the short half-life of ctDNA and RNA. For blood samples, this is complicated by the need for blood processing within a short window of time after collection. The FDA has approved the Cobas epidermal growth factor receptor (EGFR) mutation test from Roche as a ctDNA detection assay. This real-time PCR-based assay detects specific EGFR mutations in plasma and can be used to aid in the prescription of appropriate targeted therapy drugs to Non-Small Cell Lung Cancer (NSCLC) patients.
3. Sanger Sequencing: Polymerase Chain Reaction (PCR) combined with Sanger sequencing allows the identification of cancer single nucleotide variants and small insertions and deletions. The FDA has approved BRACAnalysis CDx test by Myriad, a Sanger sequencing-based test for metastatic breast, ovarian and metastatic pancreatic cancer. Although a well-established and relied-upon technique, Sanger sequencing has some limitations like any molecular assay technique. It can only address one genetic mutation at a time (single analyte approach) with a sensitivity of 15-20%, whereas NGS allows the assessment of multiple genes simultaneously (multiple analyte approach) with much higher sensitivity (down to 1%) using the same amount of input of nucleic acid.
4. Quantitative Polymerase Chain Reaction (qPCR) and Quantitative Reverse Transcription PCR (qRT-PCR): qPCR and qRT-PCR are used to detect and quantify cancer gene variants with confidence and fast turnaround. The test Therascreen EGFR RGQ by Qiagen obtained FDA-approved status for Non-Small Cell Lung Cancer (NSCLC). Quantitative PCR has the disadvantage of only allowing the interrogation of a limited set of regions of interest because of the innate reduced multiplex capability. If that is not a concern and the intention is to quantify the expression of a few genes, then this methodology is very effective. Quantitative PCR is a good option when the number of genes interrogated is low (<20 genes) and the variants are well-characterized.
5. nCounter gene expression assay: The NanoString nCounter uses capture probes and sequence-specific fluorescently labeled reporter probes to detect single-stranded RNA, DNA or protein targets in FFPE or fresh frozen samples. The unique fluorescence pattern of the reporter probe acts as molecular barcodes that can be resolved and counted during data collection, providing qualitative and quantitative data. It is possible to detect ~800 targets at once, and the introduction of biases is avoided because it does not involve enzymatic reactions (like reverse transcription and amplification). As far as FFPE RNA sample requirements for nCounter, NanoString recommends using RNA samples with DV300 of at least 50% (meaning 50% of the RNA species should be of 300 nucleotides or longer). Some RNA samples might also need an increased input of 300 ng*, which can be a rough cut-off for typical RNAs extracted from FFPE solid tumor samples.
6. Immunohistochemistry (IHC): IHC uses monoclonal and polyclonal fluorescent-dye conjugated antibodies to detect specific tumor markers (antigens) in tissue sections and cells. Although this technique is widely used and inexpensive, it is considered only semi-quantitative. PD-L1 22C3 pharmDx is an IHC test that has received FDA approval to detect the presence of programmed cell death ligand 1 (PD-L1) in head and neck squamous cell cancer, thought to be important in predicting immunotherapy response.
7. Fluorescence In Situ Hybridization (FISH): This method is capable of concomitantly localizing several genomic sequences (in the form of DNA or RNA) in cells using complementary fluorescent probes. FISH is very effective detecting copy number variation mutations (especially translocation and amplification) due to its high sensitivity and specificity. The FDA has approved a HER2 FISH test by Agilent-Dako to quantitatively determine HER2 gene amplification in breast cancer patients.
8. Hybridization capture NGS: Multiple regions of interest in the genome are captured by hybridization to biotinylated probes, isolated by magnetic pull-down and sequenced. This technique is also known as target enrichment sequencing. One of the advantages of hybridization techniques over amplification-based methods is reduced allele dropout (amplification failure of one of the two alleles at a given locus). Because the synthetic hybridization probes are longer than PCR primers, they can better tolerate the presence of mismatches in the probe binding site. Consequently, hybridization approaches are more effective in the prevention of false negatives than multiplexed PCR assays. FoundationOne CDx test as well as Memorial Sloan Kettering’s MSK-IMPACT2 test are hybridization captures approved by the FDA that use FFPE DNA to interrogate mutations in cancer genes, select gene rearrangements and microsatellite instability (MSI) and tumor mutational burden (TMB).
9. Amplicon sequencing protocols: Using a combination of PCR and NGS, it is possible to genotype solid tumors. Genomic regions known as cancer hotspots are co-amplified using multiplexed primer pools and sequenced. The FDA has approved Oncomine Dx Target Test (Thermo Fisher Scientific), a qualitative tumor profiling test for FFPE DNA from Non-Small Cell Lung Cancer (NSCLC).
10. NGS-based single-cell RNA sequencing: scRNA-seq is an emerging technique where cancer cells are isolated and sequenced individually. Tumors are a mosaic of genetically different cancer cells and immune cells that infiltrate the tumor microenvironment. Most of the genomic profile data sets thus far are obtained through the bulk sequencing of tumor tissue without the distinction between the different cell populations collected together. The expression profiles are averaged for the entire sample collected, but tumor biology is impacted by the heterogeneity of the expression of different cancer cells in the tumor. Therefore, the information obtained through scRNA-seq enables the identification of different cell types and cell states. However useful to reveal transcriptional profile heterogeneity in solid tumors, scRNA-seq assay is subject to difficulties that can lead to quantitative errors. The partial sampling of the transcriptome and sensitivity to sample quality are one of the challenges for broad adoption of scRNA-seq. Until recently this technique was restricted to fresh or fresh frozen samples, but there are recent publications indicating that laser-capture microdissection might allow FFPE samples to be processed with this method. This technique is currently being used for cancer research, and while it shows promise in cancer diagnostics, some challenges exist for adoption in the clinic at the moment.
11. Predictive Immune Modeling: While all of the technologies described above have found utility in enabling the oncology field to make critical treatment decisions, it is perhaps Next-Generation sequencing (NGS) techniques for solid tumor analysis that has truly revolutionized the clinical field. NGS has resulted in the generation of massive amounts of data that can be translated into actionable information for patient treatment. As immune-oncology drugs become more available, the need to correctly match prospective patients to these treatments increases as well. Characterizing Tumor-Infiltrating Lymphocytes (TILs) in solid tumors has shown clinical relevance to determine which patients are more likely to respond to these drugs. This is the basis for Predictive Immune Modeling, which uses machine-learning and RNA-sequencing to model the body’s immune system, quantify immune response, and combine biological signals to accurately predict patient response to therapy.
Massive amounts of RNA data generated through NGS has been harnessed by Cofactor Genomics to develop Health Expression Models, or multidimensional RNA models that represent immune cells present in the tumor microenvironment. With the ImmunoPrism assay, as low as 40 ng of FFPE RNA from solid tumors is required to quantify – with high sensitivity and specificity – the immune cells present, their cell state, and the expression levels of key genes (immune escape, co-inhibitory, co-stimulatory). Once an immune profile has been generated for each patient in a clinical study, this data can be leveraged to develop a multidimensional biomarker with appropriate patient cohorts (eg responders and non-responders to therapy). This NGS hybrid capture assay is pioneering the field of Predictive Immune Modeling for solid tumors. More information on ImmunoPrism can be found on our website and in the ImmunoPrism analytical performance validation publication.
*Tech Note “Strategies for successful gene expression assays” (2015) by Nanostring can be found in: https://www.nanostring.com/search?submit=&query=+Single+Cell+Gene+Expression+Profiling
Cofactor offers products both for Research Use Only (not to be used as a diagnostic assay) and within our CAP-certified laboratory. Please contact us to discuss which option is right for your application.