Welcome to my first blog post! In dealing with samples obtained in the clinic, as many researchers frequently do, often only a small amount of tissue is available. RNA-Seq from low input amounts had been a challenge we always wanted to address with a great solution, but when we start offering a particular service to our customers, we want to make sure that we are able to stand behind the results. For this reason, we carry out internal research projects to test our methodology. This post highlights the benefits we have obtained through our commitment to continuous testing and process improvement for our low input amount sequencing.
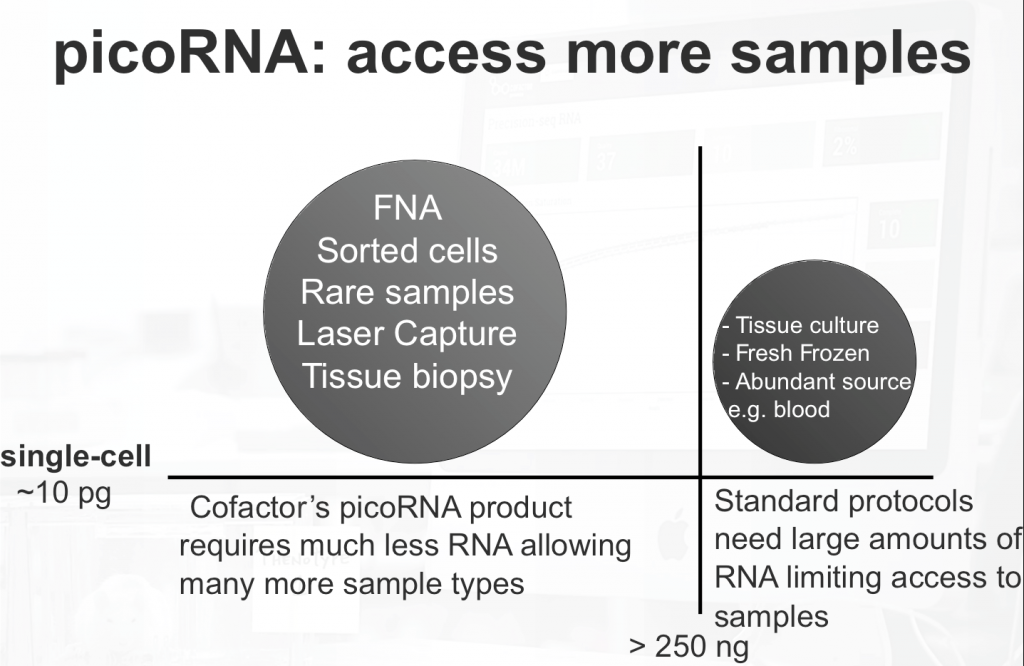
RNA-seq involving standard input amounts is becoming more routine. However, many sample types that researchers need access to fall below the amount of RNA required by most protocols. Examples of such sample sources include: fine needle aspirate, laser capture microdissection, sorted cells, tissue biopsy, and many others. Reliable, low input quantity RNA sequencing is more challenging due to the increased number of molecular manipulations (such as amplification) required to obtain a library that can be sequenced. Some of these challenges are detailed in our previous post.
Cofactor has developed a solution for low in-put RNA-seq called: picoRNA.
In order to successfully study low input samples, a scientist needs to create a library with sufficient cDNA for sequencing, to effectively remove the ribosomal RNA in the sample, and to avoid over-amplification which may lead to a lower complexity library. Though library kit manufactures include thorough explanations in the form of manuals and guides, experience is extremely useful for ensuring reliable results.
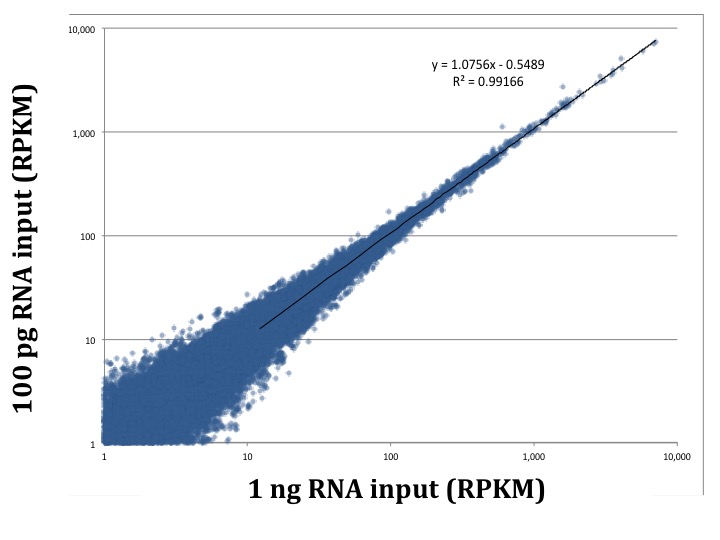
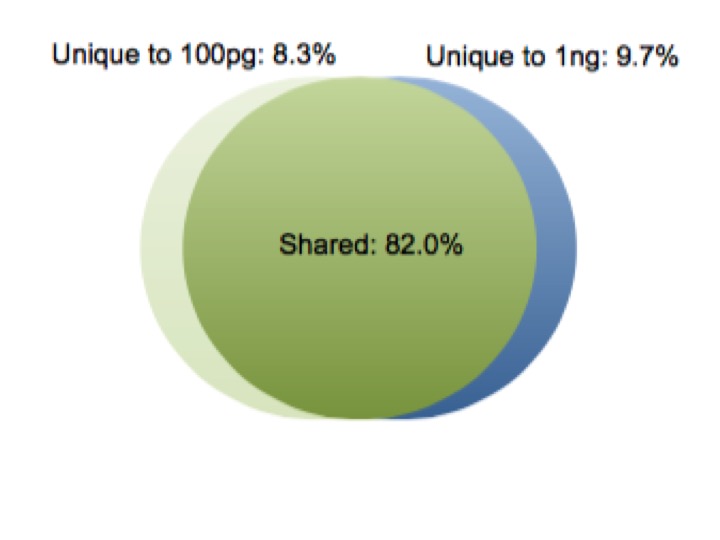
A fundamental test of a low input approach is preserving the diversity of transcripts in the sample as the amount of total RNA decreases. The earliest version of our service, picoRNA 1.0, shows consistent expression across input amounts. We compared 1 ng and 100 pg samples, and found a correlation coefficient greater than 0.99 between the two samples using picoRNA 1.0. Additionally, less than 10% of the total transcripts were unique to each input amount. As you can see in the plots below, even at a 10 fold sample input difference we are still getting consistent results. Importantly, even in the low abundance transcripts.
This opens up a large number of sample types from FNA, sorted cells, rare samples, and laser microdissection etc.
Our first iteration of picoRNA achieved consistency of transcript expression down to the 100 pg range, but we are always looking to improve our process. We want to provide the best results to our customers, and also maintain our leadership in RNA-Seq. Modifications to picoRNA were completed and further testing was required.
In order to assess our further modifications to picoRNA, I took a look at the results from six of our previous picoRNA sequencing projects (three using version 1.0, one using version 1.5 and two using version 2.0). After examining the results from all of these projects I found several metrics where our change in methodology yielded an improvement. Insert size, alignment rate and 3’ bias each showed an improvement as we modified our approach.*
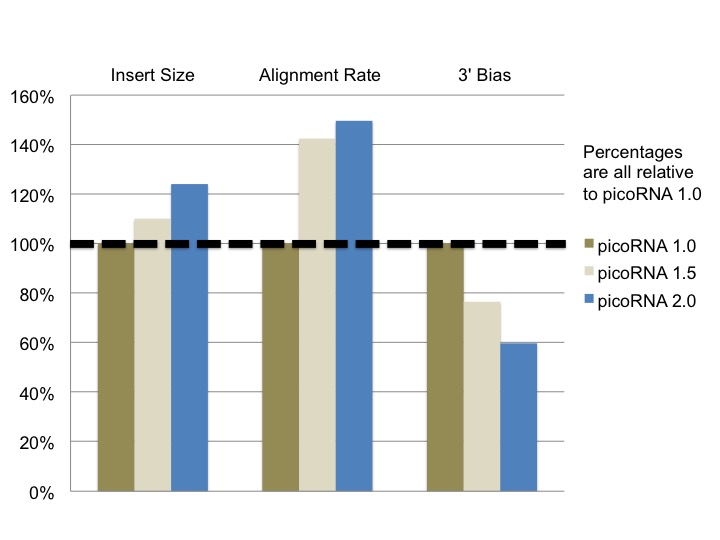
* Here 3’ bias is defined as the ratio of 3’ to 5’ ends covered of the top 1,000 expressed transcripts in a particular sample
It was great to see how our modifications ended up improving our picoRNA protocol. A greater alignment rate allows for a project to use a smaller number of raw reads, resulting in potential cost savings as not as much depth of sequencing is required. Both the longer read fragments and the more consistent, less biased coverage improves the data’s performance for isoform detection.
My favorite part of reviewing our results is seeing how Cofactor’s approach to RNA-Seq pays off. While low input kits are available to any sequencing service provider, these results show how we scrutinize our own procedures, gather data, improve our methods and deliver a superior product (in this case picoRNA). Cofactor is committed to lead the industry in sequencing RNA and improving upon the protocols that manufactures provide is one of the ways that we continue both build and utilize our knowledge.
Cofactor’s picoRNA approach enables researchers to obtain a far more complete scientific/medical picture by providing reliable results from limited material. We want to allow scientists study any samples that they may be able to obtain.