In the introduction to this series we briefly discussed tumor-infiltrating lymphocytes, or TILs. These immune cells are at the heart of immune-oncology, as they move into the tumor microenvironment and combat the tumor itself. We noted in the first piece in this series that immune cells can either help or hinder a tumor, depending on the specific immune cell and the tumor type. (For an excellent summary, see Table 1 in Fridman et al.) Thus, it remains imperative to perform immune profiling to determine how the exact immune profile of any given tumor will affect its response to therapy. Daunting, no doubt, as so many aspects of a tumor and its microenvironment change over the course of disease progression and vary from person to person.
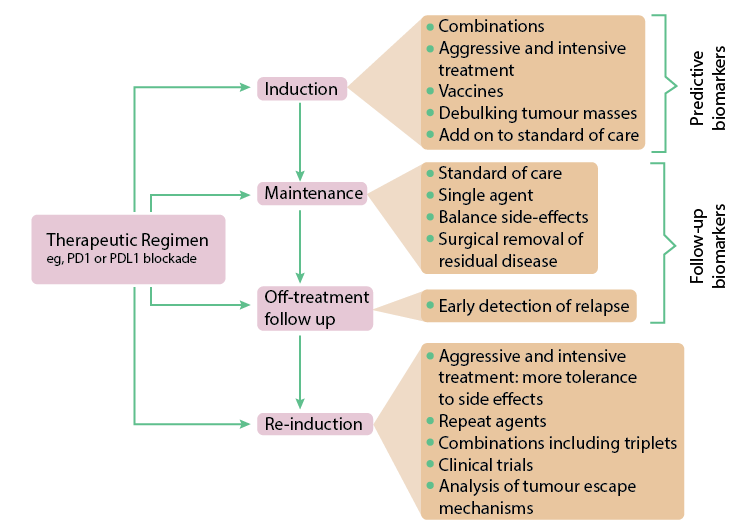
The Tumor Response
 That said, a significant body of work in immune profiling has revealed some common themes. The responsiveness of a tumor to a given treatment is now known to be influenced significantly by its immune profile. While any type of immune cell can be found in the tumor microenvironment and/or tumor itself, among the more commonly studied TILs are CD4+ and CD8+ T cells. Both proteins function in concert with the T cell receptor, helping TCR bind to MHCII and I, respectively. Thus, CD4+ and CD8+ T cells are exposed to tumor-associated antigens or neoantigens via antigen-presenting cells, allowing them to recognize and attack tumor cells and develop a memory of the antigens. Greater numbers of CD4+ and CD8+ T cells in the tumor microenvironment have been consistently linked to better prognosis. In colorectal cancer, “high densities of TILs in the primary tumor are associated with good prognosis independent of other prognostic markers” (Halama et al). That same study found that TIL density “at the invasive margin of liver metastases” was highly prognostic. More TILs predicted a better response to chemotherapy, a finding consistent with much older work such as that carried out in 1997 by Ropponen et al.
That said, a significant body of work in immune profiling has revealed some common themes. The responsiveness of a tumor to a given treatment is now known to be influenced significantly by its immune profile. While any type of immune cell can be found in the tumor microenvironment and/or tumor itself, among the more commonly studied TILs are CD4+ and CD8+ T cells. Both proteins function in concert with the T cell receptor, helping TCR bind to MHCII and I, respectively. Thus, CD4+ and CD8+ T cells are exposed to tumor-associated antigens or neoantigens via antigen-presenting cells, allowing them to recognize and attack tumor cells and develop a memory of the antigens. Greater numbers of CD4+ and CD8+ T cells in the tumor microenvironment have been consistently linked to better prognosis. In colorectal cancer, “high densities of TILs in the primary tumor are associated with good prognosis independent of other prognostic markers” (Halama et al). That same study found that TIL density “at the invasive margin of liver metastases” was highly prognostic. More TILs predicted a better response to chemotherapy, a finding consistent with much older work such as that carried out in 1997 by Ropponen et al.
On the other hand, M2 macrophages aid tumors in getting around adaptive immune responses and “promote tumor growth and progression” (Mantovani et al). This is just one example of immune cells that can support, rather than inhibit a tumor. The complexity and variability of the tumor microenvironment and immune infiltrate points to the importance of immune profiling, working to understand how individual immune cell populations, as well as the overall population, affect a tumor.
Current Methods are Flawed
The need for immune profiling is also highlighted by the fact that traditional pathology and staging can give two tumors of the same type from two different patients the same score, and yet those two patients can respond differently to treatment and end up with different outcomes. And, as Daud et al note, immunohistochemistry can indicate the presence of immune cells around a tumor but “it rarely elucidates how these cells are functioning.” Therefore, more detailed analysis of immune biomarkers is necessary and can be provided by flow cytometry, RNA-seq and microarray. Lyons et al note that “The immune contexture of the primary tumor is an essential factor in predicting prognosis. As stated above, it has been demonstrated that knowledge of the location and density of immune cells may be more prognostic than the TNM* guidelines.”
As with everything in biomedical research today, molecular level details now available through new technologies can provide deep insight into a particular medical context, as well as inform the resulting interventions. With that in mind, a group convened in 2013 to develop the “Immunoscore” that takes into consideration the level of immune infiltrate at the site of a tumor (Galon et al, 2013). Importantly, neither the Immunoscore specifically nor immune profiling in general has replaced TNM scoring. Rather, it provides a highly detailed supplement to the other factors of tumor burden, cancer cells found in lymph nodes, and metastasis. Indeed, the Immunoscore has been validated in colon cancer, and “has been used successfully to predict prognosis in melanoma, breast, kidney, and lung cancers. These results, however, have yet to be validated in prospective studies in order for the Immunoscore to be used as a predictive marker.”
Metrics like the Immunoscore are useful because they consolidate all relevant factors into a single, easily digestible number. A JNCI article from 2015 highlighted the value of the immunoscore and its validation in numerous contexts. As the field continues to move forward, the next step has been to further validate tools like this so that they can be consistently used for diagnostic, prognostic, and therapeutic purposes.
From there, it will be important to get even more granular, studying the specific immune contexture of each patient’s tumor and using it for clinical decision-making. Ultimately, this is the true promise of immune oncology.
Utilizing RNA for Immune Profiling
Fortunately, as noted above, transcriptome- and genome-level elucidation of a patient’s immune profile is now very manageable with molecular techniques such as RNA-seq. One of the great benefits to RNA-seq for immune profiling – as Lyons et al point out – is that it does not require previous knowledge of potential targets. With microarray, one builds a panel of target genes and the looks for the presence or absence of those genes in the sample. If a gene is not known to be a candidate, it won’t be included on the array and therefore cannot be detected. RNA-seq, on the other hand, allows users to approach a sample naively, discovering novel gene related to the immune response to a particular tumor.
Another challenge in immune profiling mentioned in a recent review by Duraiswamy is the limited amount of patient tissue available for analysis. RNA-seq can get around this through low-input approaches, making better use of limited resources. These approaches also hold the potential to enable more detailed assessment of possible therapeutic interventions. Lyons et al note that, “Because most non-tumor cells in the tumor microenvironment are immune cells,” the possibility for granular analysis of the immune cell population can provide “insight into the potential use of immune and other molecularly targeted therapies.” Options for targeted therapies remain limited today, but this will become more relevant as new drugs come on the market.
Even today, though, we are seeing the value of immune profiling. PD-1 is perhaps the most well know and/or most effective target for immune therapy. Even a brief review of the literature brings this molecule to the forefront. For example, just over a year ago, Daud et al published a study in the Journal of Clinical Investigation titled, “Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma.” As with all good publications, the title clearly states the primary finding. More specifically, though, it is the level of CD8+ T cells expressing PD-1 that are correlated with therapeutic response. Ahmadzadeh et al demonstrated that TILs expressing PD-1 are functionally impaired and have an “exhausted” phenotype that limits their ability to respond to the tumor.
Interestingly, the recent focus on PD-1 as a target has led to some controversy. According to Madan and Gulley, simply staining for PD-1 and it’s ligand PD-L1 doesn’t provide enough resolution and raises questions about “the threshold for a ‘positive’ finding.” Put another way, the debate becomes one of somewhat subjective histological analysis. Furthermore, even PD-1/PD-L1 positive biopsies, only about
1/5 respond to anti-PD-1 therapy. The reason is that, once again, the tug-of-war between the tumor and immune system causes variability in the underlying immune profile at the microenvironment. With additional immune profiling, there is the possibility to utilize combination therapies to better activate an immune response and obtain positive outcomes in patients who do not respond to anti-PD-1 treatment.
The good news is that, like so much in science and medicine, the field has advanced rapidly over the past decade. Indeed, a 2007 review in Cancer Research by Galan et al stated that “[G]enotypes, as well as some molecular, protein, and carbohydrate markers, have been shown to influence clinical outcome. None of these factors have been validated yet as robust independent prognostic markers for patient care” (emphasis added). Today, as noted above, immune profiling has become a valuable component in diagnosis and prognosis. Furthermore, the genotyping described in that paper consisted of DNA microarray. Since then, RNA-seq has largely supplanted microarray for gene profiling.
So, the tools exist to perform robust immune profiling of any given tumor. The challenge going forward will be to create and evaluate therapies that can take advantage of this information (which really describes the challenge behind the entire precision medicine movement!)
Predictive Immune Modeling: A New Discipline
 The ImmunoPrism™ assay has been designed with these immune profiling goals in mind. It leverages Cofactor’s Health Expression Models, which were built to capture a new way of looking at biology — with greater resolution and depth than isolated, single-analyte markers. These multidimensional, gene expression models are pathway-agnostic and rely on the dynamic nature of RNA to define what makes a patient, cell type, or disease unique.
The ImmunoPrism™ assay has been designed with these immune profiling goals in mind. It leverages Cofactor’s Health Expression Models, which were built to capture a new way of looking at biology — with greater resolution and depth than isolated, single-analyte markers. These multidimensional, gene expression models are pathway-agnostic and rely on the dynamic nature of RNA to define what makes a patient, cell type, or disease unique.
When working with patient cohorts from retrospective studies with clinical outcomes, these individual immune profiles are leveraged using Cofactor’s Predictive Immune Modeling platform. They help identify the most powerful combination of analytes to generate a multidimensional biomarker. The predictive accuracy of this multidimensional biomarker is presented in the Biomarker Report, which includes statistics, selection thresholds, and more. Contact us at [email protected] for more information about immune profiling in cancer and how the ImmunoPrism assay could be used in your work.
<<Previous Article Next Article>>
*Note: TNM scoring refers to the traditional method of grading solid tumors based on three criteria: T, Primary tumor size; N, regional lymph node involvement; M, distal metastases.

