How is Cofactor’s RNA-seq different?
How is Cofactor’s RNA-seq different?
Studying RNA gives researchers a powerful insight into biological differences and genes and pathways that may be associated with interesting biological characteristics. These studies are however not perfect. For each RNA study, researchers must address variable sample quality, unknown sample complexity, and provide confident yet easy results navigation to be able to unlock the power of the technology.
Cofactor’s RNA-seq offering has been developed to overcome these limitations by increasing the sensitivity, specificity, and accuracy of your RNA-seq experiments. Cofactor’s RNA-seq gives you information on the quality of each step of the experiment, helps you understand when to stop sequencing, compares complexity across samples and presents a place to start finding answers. Below are some of the key ways that Cofactor’s RNA-seq provides bioinformaticians, biologists, or even non scientific managers the most insightful molecular and data assessment and characterization of anyone in the industry. This translates to more consistent results and faster answers to your scientific questions.
Spike-in Controls
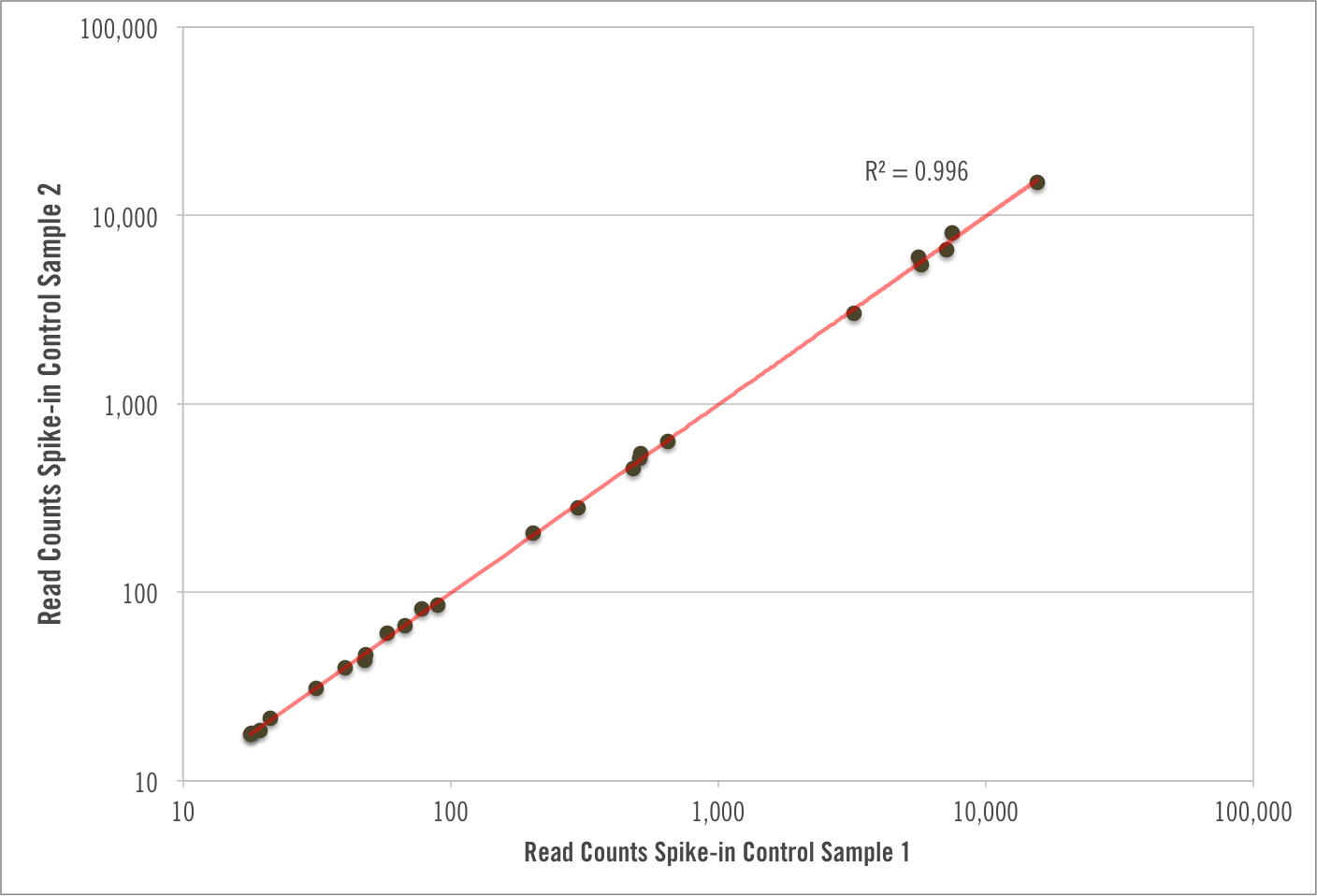
Historically there has been a lack of confidence in RNA-seq studies relating to the large number of molecular manipulations. Slight differences in procedure can sometimes have a large impact on expression between samples.
Cofactor’s RNA-seq gets rid of the ambiguity by incorporating synthetic spike-in controls from the very first step. Spike-ins provide quality assurance and enable you to disambiguate true biological differences from process variability.
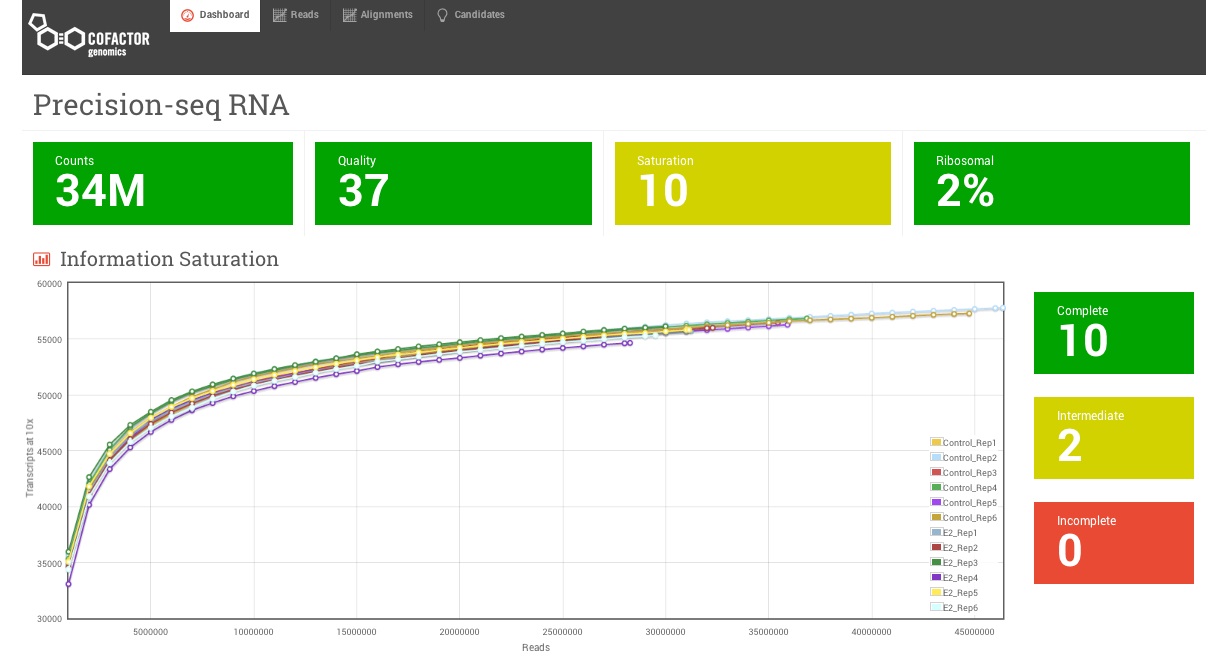
Saturation Curves
In many cases, researchers guess at how many sequencing reads are required. With saturation curves you don’t need to guess.
Cofactor’s RNA-seq generates a saturation report for every sample, providing you insight and confidence in how your samples performed. You’ll understand how many reads you need, how many genes are expressed in your samples, and whether there are questionable samples that should be excluded from comparison. This information is powerful for pilot studies prior to ramp up, and helps identify outlier or low complexity samples to be excluded from analysis in larger studies.
Cofactor’s ActiveSite
One of the largest hurdles to RNA-seq has been an intuitive interface that puts the power of discovery in the hands of those most involved with the goals of the project be they the bioinformatician, biologist, or project director.
ActiveSite provides a web-based interface for discovery. It starts with an overview of how your project scored on important quality metrics, and then leads to a discovery interface designed to help you navigate and sort through RNA-seq data. It offers simplicity and ease of use on the surface for quick browsing, while still letting you dive deep in to the data when you need to.
Learn more about ActiveSite.
Don’t settle. Learn why top researchers choose Cofactor.
Get a copy of our ebook: “Six Habits of Highly Successful RNA-seq Researchers” today.




